Decoding Hidden Messages with the Central Dogma
Abstract
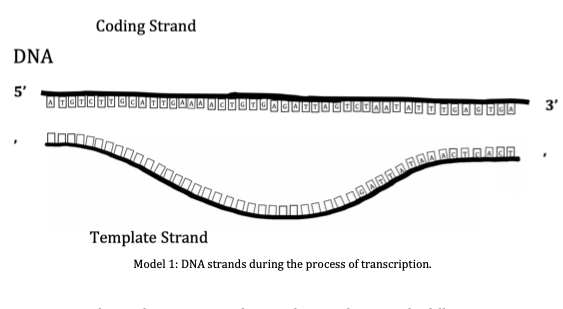
In “Decoding Hidden Messages with the Central Dogma" students explore distinct DNA, RNA and Ribosome models to predict base pairing, decode hidden messages within nucleotide sequences, define RNA polymerase and ribosome directionality. Students utilize amino acid models and charts to apply the triplet base code in the formation of a peptide chain throughout translation. Furthermore, students compare non-mutated sequences with mutated sequences and learn to define four distinct types of mutations – Silent, missense, nonsense, and frameshift.
The content objectives of this activity are: 1. Prediction of directionality of RNA polymerase during transcription and ribosomes during translation. 2. Application of the triplet base code to decode base sequence messages. 3. Definition of distinct types of mutations within sequences. The Process objectives in this activity are: 1. Student engagement in group work based in distinct participation roles. 2. Application of self-assessment methods. 3. Sharing of knowledge within and beyond student groups.
Level: Undergraduate
Setting: Classroom
Activity Type: Learning Cycle, Application
Discipline: General Biology
Course: Introduction to Biology
Keywords: Central Dogma, DNA, RNA, Transcription, Translation, Mutations
Downloads
Published
How to Cite
Issue
Section
License
Copyright of this work and the permissions granted to users of the PAC are defined in the PAC Activity User License.

